measure allowing to build an histogram of disparity between angle measured on shape and angle measured on reference ellipse
This measure allows to quantified the angularity of a shape using the difference between angle of consecutive segments of polygonal approximation of shape with angle on its equivalent ellipse.
Note that equivalent ellipse associated to a shape is define such as :
- ellipse center is set at shape barycenter (see Barycenter2d)
- ellipse orientation is equal to orientation extracted from second order moment of shape (see Inertia Orientation 2d)
- ellipse dimensions are defined such that ellipse perimeter and area match with shape perimeter (see Perimeter2d) and shape area (see Area2d).
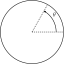
For each tops of the polygonal approximation of shape, we compute an angularity value given by :
![\[ A_i = \left |\theta_i - \phi_i \right | \]](form_1007.png)
where :
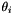 is angle between segments ending at considered top
is angle between segments ending at considered top 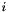 .
.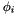 is the associated angle on equivalent ellipse.
is the associated angle on equivalent ellipse.
We then compute the histogram of these values using a number of orientation defined by parameter NbOrientations.
The global shape angularity value is then given by mean value associated to bin containing quantile value defined by parameter Quantile.
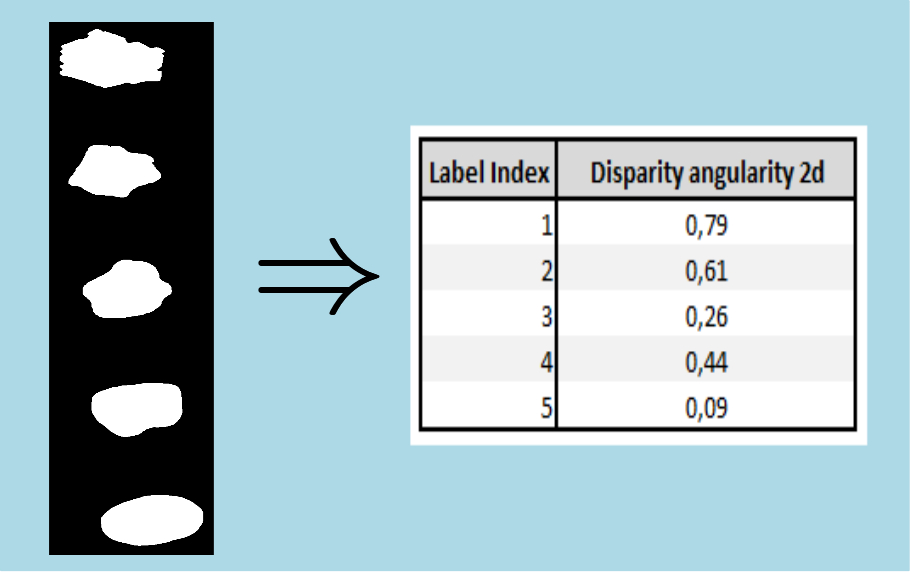
Here is an example of degree of angularity 2d computation on few caracteristic shapes :
measure allowing to build an histogram of disparity between angle measured on shape and angle measured on reference ellipse
Measure synthesis :
| Measure Type | Measure Unit Type | Parameter Type | Result Type | Shape Requirements |
Geometry 2d
|
Angle
|
DisparityAngularity2dMsrParams |
Value (ipsdk::ipReal64)
|
Both
|
See Shape measurement for additional information on these pictograms Example of Python code :
Generic example in 2d case :
import PyIPSDK
import PyIPSDK.IPSDKIPLShapeAnalysis as shapeanalysis
inMeasureInfoSet2d = PyIPSDK.createMeasureInfoSet2d()
PyIPSDK.createMeasureInfo(inMeasureInfoSet2d, "GradientAngularityIndex2dMsr")
outMeasureSet = shapeanalysis.labelAnalysis2d(inGreyImg, inLabelImg2d, inMeasureInfoSet2d)
PyIPSDK.saveCsvMeasureFile(os.path.join(tmpPath, "shape_analysis_results.csv"), outMeasureSet)
outMsr = outMeasureSet.getMeasure("GradientAngularityIndex2dMsr")
outMsrValues = outMsr.getMeasureResult().getColl(0)
print("First label measurement equal " + str(outMsrValues[1]))
Example of C++ code :
Example informations
Associated library
IPSDKIPLShapeAnalysis
Code Example
Shape2dCollPtr pShape2dColl = boost::make_shared<Shape2dColl>();
IPSDK_REQUIRE(readFromXmlFile(inputShape2dCollPath, *pShape2dColl) == true);
MeasureInfoSetPtr pMeasureInfoSet = MeasureInfoSet::create2dInstance();
createMeasureInfo(pMeasureInfoSet, "DisparityAngularity2dMsr");
MeasureSetPtr pOutMeasureSet =
shapeAnalysis2d(pInGreyImg2d, pShape2dColl, pMeasureInfoSet);
const MeasureConstPtr& pDisparityAngularity2dOutMsr = pOutMeasureSet->getMeasure("DisparityAngularity2dMsr");
const ipsdk::shape::analysis::ValueMeasureResult<ipsdk::ipReal64>& outResults = static_cast<const ipsdk::shape::analysis::ValueMeasureResult<ipsdk::ipReal64>&>(pDisparityAngularity2dOutMsr->getMeasureResult());

![\[ A_i = \left |\theta_i - \phi_i \right | \]](form_1007.png)
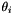 is angle between segments ending at considered top
is angle between segments ending at considered top 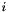 .
.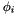 is the associated angle on equivalent ellipse.
is the associated angle on equivalent ellipse. 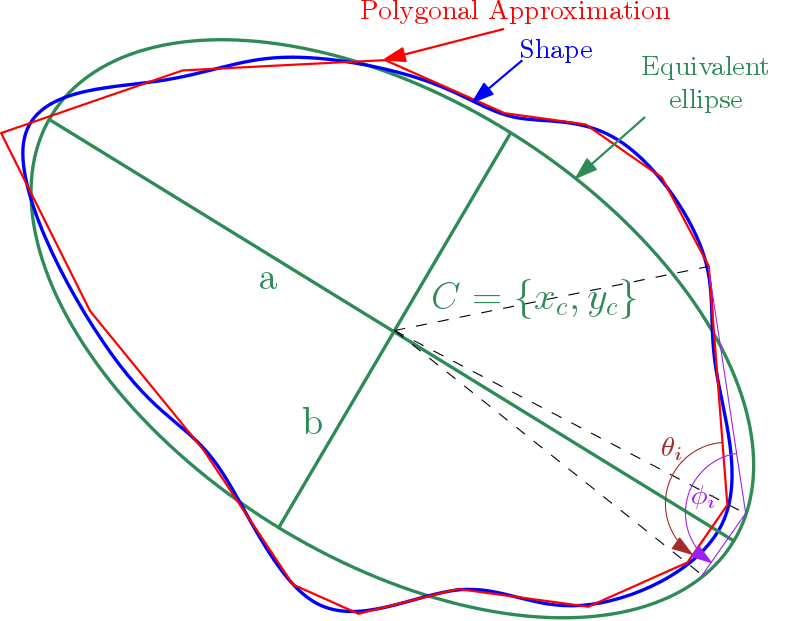




 1.8.14
1.8.14