 |
IPSDK 4.1.1.1
IPSDK : Image Processing Software Development Kit
|
Measure allowing to compute energy of intensities for shape.
This measure computes the energy, also known as angular second moment, of image pixel/voxel intensity values associated to a 2d/3d shape. Its value equals to one for a constant signal and decreases with its unpredictability.
The energy measure is based on the analysis of the shape histogram (see Histogram). Given the histogram parameters allowing to determine (among others) the number of classes 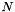 and a global range for the histogram, this measure computes shape energy as :
and a global range for the histogram, this measure computes shape energy as :
![\[ E = \sum_{i=0}^{N-1}{p^2(b_i)} \]](form_1119.png)
where 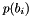 is the density of probability associated to bin
is the density of probability associated to bin 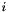 of histogram :
of histogram :
![\[ p(b_i) = \frac{P(b_i) \times R(b_i)}{\sum_{i=0}^{N-1}{P(b_i) \times R(b_i)}} \]](form_1121.png)
and :
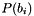 is the population for the bin
is the population for the bin 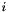 of histogram,
of histogram,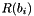 is the intensity range (width) for the bin
is the intensity range (width) for the bin 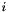 of histogram.
of histogram.Here is an example of energy measurement in 2d case :
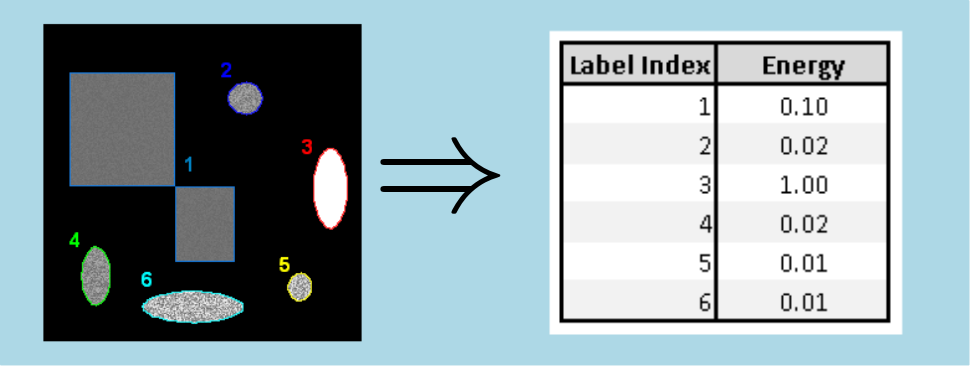
We could have predicted the behaviour of the results : the shape 3 has homogeneous intensity, thus, its energy equals 1. Moreover, the shapes 5 and 6 are the most noisy features, therefore their energies have the lowest values. Then, the noise intensity of the shapes 2 and 4, is close to the intensity noise intensity of the shapes 5 and 6. As a consequence, their energy have the same range. Finally, the shape 1 is less noisy than the shapes 2, 4, 5 and 6, which results in a higher energy value.
Moreover, the results are correlated with the ones obtained by the Entropy measure.
Measure allowing to compute energy of intensities for shape
| Measure Type | Measure Unit Type | Parameter Type | Result Type | Shape Requirements |
|---|---|---|---|---|

Generic | 
None | 
| 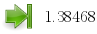
Value (ipsdk::ipReal64) | 
Row Intersections |
Generic example in 2d case :
Generic example in 3d case :
IPSDKIPLShapeAnalysis
 1.8.14
1.8.14