| image = | normalizedCrossCorrelation3dImg (inImg3d,inKnlXYZ) |
| image = | normalizedCrossCorrelation3dImg (inImg3d,inKnlImg3d) |
Computes the Normalized Cross Correlation between a volume and a 3d kernel.
The three dimensional Normalized Cross-Correlation (NCC) between a volume 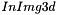 and a template
and a template 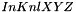 is the 3d extension of the 2d Normalized Cross-Correlation. It is defined in spatial domain as follows :
is the 3d extension of the 2d Normalized Cross-Correlation. It is defined in spatial domain as follows :
![\[ NCC(InImg3d(\textbf{x}), InKnlXYZ) = \frac{\sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}}{ \left( InImg3d(\textbf{x}) - \overline{InImg3d} \right) \left( InKnlXYZ(\textbf{x}+\textbf{i}) - \overline{InKnlXYZ} \right) }} {\sqrt{ \sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}} { \left( InImg3d(\textbf{x}) - \overline{InImg3d} \right)^2} \sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}} { \left( InKnlXYZ(\textbf{x}+\textbf{i}) - \overline{InKnlXYZ} \right)^2}}} \]](form_644.png)
Where ![$ \textbf{x} = \left[ x, y, z \right]^T $](form_645.png) represents the coordinates of a voxel,
represents the coordinates of a voxel, 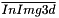 is the mean of
is the mean of 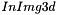 in the neighbourhood defined by the kernel and
in the neighbourhood defined by the kernel and 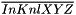 is the mean of
is the mean of 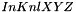 .
.
See Normalized Cross-Correlation 2d for an illustration of a 2d normalized cross-correlation result.
- See also
- http://en.wikipedia.org/wiki/Cross-correlation
- Note
- It is possible to call the NCC with an image provided as a template, nevertheless the algorithm still internally calculates and uses a kernel. These functions are ment to simplify the use of the function but it means that the data are copied in memory.
Example of Python code :
Example imports
import PyIPSDK
import PyIPSDK.IPSDKIPLFiltering as filter
Code Example
inImg = PyIPSDK.loadTiffImageFile(inputImgPath)
inKnl = PyIPSDK.rectangularKernelXYZ(1, 2, 1, [1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 0, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1,
1, 1, 1],
True)
outImg = filter.normalizedCrossCorrelation3dImg(inImg, inKnl)
Example of C++ code :
Example informations
Header file
#include <IPSDKIPL/IPSDKIPLFiltering/Processor/NormalizedCrossCorrelation3dImg/NormalizedCrossCorrelation3dImg.h>
Code Example
ImagePtr pOutNCCImg = normalizedCrossCorrelation3dImg(pImg, pKernel);

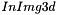 and a template
and a template 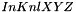 is the 3d extension of the 2d Normalized Cross-Correlation. It is defined in spatial domain as follows :
is the 3d extension of the 2d Normalized Cross-Correlation. It is defined in spatial domain as follows :![\[ NCC(InImg3d(\textbf{x}), InKnlXYZ) = \frac{\sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}}{ \left( InImg3d(\textbf{x}) - \overline{InImg3d} \right) \left( InKnlXYZ(\textbf{x}+\textbf{i}) - \overline{InKnlXYZ} \right) }} {\sqrt{ \sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}} { \left( InImg3d(\textbf{x}) - \overline{InImg3d} \right)^2} \sum \limits_{\textbf{i $ \in \aleph_{\textbf{x}}$}} { \left( InKnlXYZ(\textbf{x}+\textbf{i}) - \overline{InKnlXYZ} \right)^2}}} \]](form_644.png)
![$ \textbf{x} = \left[ x, y, z \right]^T $](form_645.png) represents the coordinates of a voxel,
represents the coordinates of a voxel, 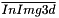 is the mean of
is the mean of 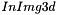 in the neighbourhood defined by the kernel and
in the neighbourhood defined by the kernel and 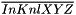 is the mean of
is the mean of 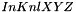 .
.